Bayesian Population Analysis using WinBUGSのコードをStanに移植してみる(第7章その4) [統計]
第7章その3につづいて、第7章の残りの部分です。CJSモデルを、m-arrayをつかって多項尤度で定式化しています。BUGSでは、多項尤度の方が状態空間モデルよりも収束がはやいと本文にありますが、Stanでも同様です。
7.10. Fitting the CJS to data in the m-array format: the multinomial likelihood
7.10.2. Time-dependent models
cjs_mnl_code <- "
data {
int<lower=0> n_occasions;
int<lower=0> marr[n_occasions-1,n_occasions];
}
transformed data {
int r[n_occasions-1];
// Calculate the number of birds released each year
for (t in 1:(n_occasions - 1))
r[t] <- sum(marr[t]);
}
parameters {
vector<lower=0,upper=1>[n_occasions - 1] phi;
vector<lower=0,upper=1>[n_occasions - 1] p;
}
transformed parameters {
vector<lower=0,upper=1>[n_occasions - 1] q;
simplex[n_occasions] pr[n_occasions - 1];
q <- 1.0 - p; // Probability of non-recapture
// Define the cell probabilities of the m-array
// Main diagonal
for (t in 1:(n_occasions - 1)) {
pr[t][t] <- phi[t] * p[t];
// Above main diagonal
for (j in (t + 1):(n_occasions - 1)) {
pr[t][j] <- prod(segment(phi, t, j - t + 1)) *
prod(segment(q, t, j - t)) *
p[j];
} //j
// Below main diagonal
for (j in 1:(t - 1))
pr[t][j] <- 0;
} //t
// Last column: probability of non-recapture
for (t in 1:(n_occasions - 1))
pr[t][n_occasions] <- 1 - sum(head(pr[t], n_occasions - 1));
}
model {
// Priors
phi ~ uniform(0, 1); // Priors for survival
p ~ uniform(0, 1); // Priors for recapture
// Define the multinomial likelihood
for (t in 1:(n_occasions - 1)) {
increment_log_prob(multinomial_log(marr[t], pr[t]));
}
}
generated quantities {
real fit;
real fit_new;
matrix[n_occasions - 1, n_occasions] E_org;
matrix[n_occasions - 1, n_occasions] E_new;
vector[n_occasions] expmarr[n_occasions - 1];
int<lower=0> marr_new[n_occasions - 1, n_occasions];
// Assess model fit using Freeman-Tukey statistic
// Compute fit statistics for observed data
for (t in 1:(n_occasions - 1)){
expmarr[t] <- r[t] * pr[t];
for (j in 1:n_occasions){
E_org[t, j] <- square((sqrt(marr[t][j]) - sqrt(expmarr[t][j])));
} //j
} //t
// Generate replicate data and compute fit stats from them
for (t in 1:(n_occasions - 1)) {
marr_new[t] <- multinomial_rng(pr[t], r[t]);
for (j in 1:n_occasions) {
E_new[t, j] <- square((sqrt(marr_new[t][j]) - sqrt(expmarr[t][j])));
}
}
fit <- sum(E_org);
fit_new <- sum(E_new);
}
"
## Create the m-array from the capture-histories
marr <- marray(CH)
## Bundle data
stan_data <- list(marr = marr, n_occasions = dim(marr)[2])
# Parameters monitored
params <- c("phi", "p", "fit", "fit_new")
# MCMC settings
ni <- 2000
nt <- 1
nb <- 1000
nc <- 3
## Initial values
inits <- lapply(1:nc, function(i) list(phi = runif(dim(marr)[2]-1, 0, 1),
p = runif(dim(marr)[2]-1, 0, 1)))
## Stan
model <- stan_model(model_code = cjs_mnl_code)
cjs <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
7.10.3. Age-dependent models
cjs_mnl_age_code <- "
data {
int<lower=0> n_occasions;
int<lower=0> marr_j[n_occasions - 1, n_occasions];
int<lower=0> marr_a[n_occasions - 1, n_occasions];
}
parameters {
real<lower=0,upper=1> mean_phijuv;
real<lower=0,upper=1> mean_phiad;
real<lower=0,upper=1> mean_p;
}
transformed parameters {
vector<lower=0,upper=1>[n_occasions - 1] phi_juv;
vector<lower=0,upper=1>[n_occasions - 1] phi_ad;
vector<lower=0,upper=1>[n_occasions - 1] p;
vector<lower=0,upper=1>[n_occasions - 1] q;
simplex[n_occasions] pr_j[n_occasions - 1];
simplex[n_occasions] pr_a[n_occasions - 1];
// Constraints
for (t in 1:(n_occasions - 1)) {
phi_juv[t] <- mean_phijuv;
phi_ad[t] <- mean_phiad;
p[t] <- mean_p;
}
q <- 1.0 - p; // Probability of non-recapture
// Define the cell probabilities of the m-arrays
// Main diagonal
for (t in 1:(n_occasions - 1)) {
pr_j[t][t] <- phi_juv[t] * p[t];
pr_a[t][t] <- phi_ad[t] * p[t];
// Above main diagonal
for (j in (t + 1):(n_occasions - 1)) {
pr_j[t][j] <- phi_juv[t] *
prod(segment(phi_ad, t + 1, j - t)) *
prod(segment(q, t, j - t)) *
p[j];
pr_a[t][j] <- prod(segment(phi_ad, t, j - t + 1)) *
prod(segment(q, t, j - t)) *
p[j];
} // j
// Below main diagonal
for (j in 1:(t - 1)) {
pr_j[t][j] <- 0;
pr_a[t][j] <- 0;
} //j
} // t
// Last column: probability of non-recapture
for (t in 1:(n_occasions - 1)) {
pr_j[t][n_occasions] <- 1 - sum(head(pr_j[t], n_occasions - 1));
pr_a[t][n_occasions] <- 1 - sum(head(pr_a[t], n_occasions - 1));
} // t
}
model {
// Priors
mean_phijuv ~ uniform(0, 1); // Prior for mean juv. survival
mean_phiad ~ uniform(0, 1); // Prior for mean ad. survival
mean_p ~ uniform(0, 1); // Prior for mean recapture
// Define the multinomial likelihood
for (t in 1:(n_occasions - 1)) {
increment_log_prob(multinomial_log(marr_j[t], pr_j[t]));
increment_log_prob(multinomial_log(marr_a[t], pr_a[t]));
}
}
"
## Bundle data
stan_data <- list(marr_j = CH.J.marray,
marr_a = CH.A.marray,
n_occasions = dim(CH.J.marray)[2])
## Initial values
inits <- function(){list(mean_phijuv = runif(1, 0, 1),
mean_phiad = runif(1, 0, 1),
mean_p = runif(1, 0, 1))}
## Parameters monitored
params <- c("mean_phijuv", "mean_phiad", "mean_p")
# MCMC settings
ni <- 2000
nt <- 1
nb <- 1000
nc <- 3
## Stan
model <- stan_model(model_code = cjs_mnl_age_code)
cjs_2 <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
7.11. Analysis of a real data set: survival of female Leisler’s bats
cjs_mnl_ran_code <- "
data {
int<lower=0> n_occasions;
int<lower=0> marr[n_occasions - 1, n_occasions];
}
transformed data {
int r[n_occasions];
for (t in 1:n_occasions - 1)
r[t] <- sum(marr[t]);
}
parameters {
real<lower=0,upper=1> mean_phi;
real<lower=0,upper=1> mean_p;
real<lower=0> sigma;
real epsilon[n_occasions - 1];
}
transformed parameters {
real mu;
vector<lower=0,upper=1>[n_occasions - 1] phi;
vector<lower=0,upper=1>[n_occasions - 1] p;
vector<lower=0,upper=1>[n_occasions - 1] q;
simplex[n_occasions] pr[n_occasions - 1];
mu <- logit(mean_phi); // Logit transformation
// Constraints
for (t in 1:(n_occasions - 1)) {
phi[t] <- inv_logit(mu + epsilon[t]);
p[t] <- mean_p;
}
q <- 1.0 - p; // Probability of non-recapture
// Define the cell probabilities of the m-arrays
// Main diagonal
for (t in 1:(n_occasions - 1)) {
pr[t][t] <- phi[t] * p[t];
// Above main diagonal
for (j in (t + 1):(n_occasions - 1))
pr[t][j] <- prod(segment(phi, t, j - t + 1)) *
prod(segment(q, t, j - t)) *
p[j];
// Below main diagonal
for (j in 1:(t - 1))
pr[t][j] <- 0;
} // t
// Last column: probability of non-recapture
for (t in 1:(n_occasions - 1))
pr[t][n_occasions] <- 1 - sum(head(pr[t], n_occasions - 1));
}
model {
// Priors
epsilon ~ normal(0, sigma);
mean_phi ~ uniform(0, 1); // Prior for mean juv. survival
sigma ~ uniform(0, 5);
mean_p ~ uniform(0, 1); // Prior for mean recapture
// Define the multinomial likelihood
for (t in 1:(n_occasions - 1))
increment_log_prob(multinomial_log(marr[t], pr[t]));
}
generated quantities {
real<lower=0> sigma2;
real<lower=0> sigma2_real;
vector[n_occasions] expmarr[n_occasions - 1];
int marr_new[n_occasions - 1, n_occasions];
matrix[n_occasions - 1, n_occasions] E_org;
matrix[n_occasions - 1, n_occasions] E_new;
real fit;
real fit_new;
sigma2 <- square(sigma);
// Temporal variance on real scale
sigma2_real <- sigma2 * square(mean_phi) * square(1 - mean_phi);
// Assess model fit using Freeman-Tukey statistic
// Compute fit statistics for observed data
for (t in 1:(n_occasions - 1)) {
expmarr[t] <- r[t] * pr[t];
for (j in 1:n_occasions)
E_org[t, j] <- square((sqrt(marr[t, j]) - sqrt(expmarr[t][j])));
}
// Generate replicate data and compute fit stats from them
for (t in 1:(n_occasions - 1)) {
marr_new[t] <- multinomial_rng(pr[t], r[t]);
for (j in 1:n_occasions)
E_new[t, j] <- square((sqrt(marr_new[t, j]) - sqrt(expmarr[t][j])));
}
fit <- sum(E_org);
fit_new <- sum(E_new);
}
"
## Bundle data
stan_data <- list(marr = m.leisleri,
n_occasions = dim(m.leisleri)[2])
## Initial values
inits <- function(){list(mean_phi = runif(1, 0, 1),
sigma = runif(1, 0, 5),
mean_p = runif(1, 0, 1))}
## Parameters monitored
params <- c("phi", "mean_p", "mean_phi", "sigma2", "sigma2_real",
"fit", "fit_new")
# MCMC settings
ni <- 3000
nt <- 2
nb <- 1000
nc <- 4
## Stan
model <- stan_model(model_code = cjs_mnl_ran_code)
leis_result <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
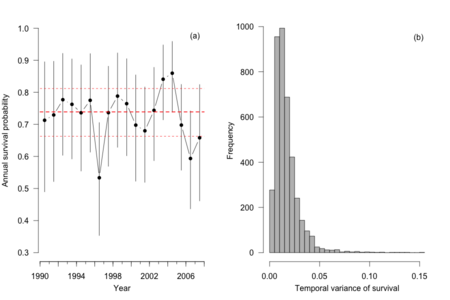
結果の図(P.235)を出力するRコードは以下のようになります。
## Produce figure of female survival probabilities
post_samp <- extract(leis_result)
post_mean <- get_posterior_mean(leis_result)[, "mean-all chains"];
par(mfrow = c(1, 2), las = 1, mar=c(4, 4, 2, 2), mgp = c(3, 1, 0))
lower <- upper <- numeric()
T <- dim(m.leisleri)[2]-1
for (t in 1:T){
lower[t] <- quantile(post_samp$phi[, t], 0.025)
upper[t] <- quantile(post_samp$phi[, t], 0.975)
}
plot(y = post_mean[grep("^phi", names(post_mean))],
x = (1:T) + 0.5, type = "b", pch = 16,
ylim = c(0.3, 1), ylab = "Annual survival probability",
xlab = "", axes = F)
axis(1, at = seq(1,(T + 1), 2), labels = seq(1990, 2008, 2))
axis(1, at = 1:(T + 1), labels = rep("", T + 1), tcl = -0.25)
axis(2, las = 1)
mtext("Year", 1, line = 2.25)
text(18, 0.975, "(a)")
segments((1:T) + 0.5, lower, (1:T) + 0.5, upper)
m <- post_mean["mean_phi"]
l <- quantile(post_samp$mean_phi, 0.025)
u <- quantile(post_samp$mean_phi, 0.975)
segments(1, m, T + 1, m, lty = 2, col = "red", lwd = 2)
segments(1, l, T + 1, l, lty = 2, col = "red")
segments(1, u, T + 1, u, lty = 2, col = "red")
hist(post_samp$sigma2_real, nclass = 45, col = "gray",
main = "", las = 1, xlab = "")
mtext("Temporal variance of survival", 1, line = 2.25)
text(max(post_samp$sigma2_real), 950, "(b)", adj = 1)
タグ:STAn





コメント 0