Bayesian Population Analysis using WinBUGSのコードをStanに移植してみる(第7章その3) [統計]
第7章その2のつづきです。
7.7. Models with age effects
cjs_age_code <- "
functions {
int first_capture(int[] y_i) {
for (k in 1:size(y_i))
if (y_i[k])
return k;
return 0;
}
int last_capture(int[] y_i) {
for (k_rev in 0:(size(y_i) - 1)) {
int k;
k <- size(y_i) - k_rev;
if (y_i[k])
return k;
}
return 0;
}
matrix prob_uncaptured(int nind, int n_occasions,
matrix p, matrix phi) {
matrix[nind, n_occasions] chi;
for (i in 1:nind) {
chi[i, n_occasions] <- 1.0;
for (t in 1:(n_occasions - 1)) {
int t_curr;
int t_next;
t_curr <- n_occasions - t;
t_next <- t_curr + 1;
chi[i, t_curr] <- (1 - phi[i, t_curr]) +
phi[i, t_curr] *
(1 - p[i, t_next - 1]) *
chi[i, t_next];
}
}
return chi;
}
}
data {
int<lower=0> nind;
int<lower=2> n_occasions;
int<lower=0,upper=1> y[nind, n_occasions];
int<lower=1> max_age;
int<lower=0,upper=max_age> x[nind, n_occasions - 1];
}
transformed data {
int<lower=0,upper=n_occasions> first[nind];
int<lower=0,upper=n_occasions> last[nind];
for (i in 1:nind)
first[i] <- first_capture(y[i]);
for (i in 1:nind)
last[i] <- last_capture(y[i]);
}
parameters {
real<lower=0,upper=1> mean_phi;
real<lower=0,upper=1> mean_p;
real<lower=0,upper=1> beta[max_age];
}
transformed parameters {
matrix<lower=0,upper=1>[nind, n_occasions - 1] phi;
matrix<lower=0,upper=1>[nind, n_occasions - 1] p;
matrix<lower=0,upper=1>[nind, n_occasions] chi;
// Constraints
for (i in 1:nind) {
for (t in 1:(first[i] - 1)) {
phi[i, t] <- 0;
p[i, t] <- 0;
}
for (t in first[i]:(n_occasions - 1)) {
phi[i, t] <- beta[x[i, t]];
p[i, t] <- mean_p;
}
}
chi <- prob_uncaptured(nind, n_occasions, p, phi);
}
model {
// Priors
beta ~ uniform(0, 1); // Prior for mean survival
mean_p ~ uniform(0, 1); // Prior for mean recapture
// Likelihood
for (i in 1:nind) {
if (first[i] > 0) {
for (t in (first[i] + 1):last[i]) {
1 ~ bernoulli(phi[i, t - 1]);
y[i, t] ~ bernoulli(p[i, t - 1]);
}
1 ~ bernoulli(chi[i, last[i]]);
}
}
}
"
## Bundle data
## Create matrices X indicating age classes
## NA -> 0
x.j <- matrix(0, ncol = dim(CH.J)[2]-1, nrow = dim(CH.J)[1])
x.a <- matrix(0, ncol = dim(CH.A)[2]-1, nrow = dim(CH.A)[1])
for (i in 1:dim(CH.J)[1]){
for (t in f.j[i]:(dim(CH.J)[2]-1)){
x.j[i,t] <- 2
x.j[i,f.j[i]] <- 1
} #t
} #i
for (i in 1:dim(CH.A)[1]){
for (t in f.a[i]:(dim(CH.A)[2]-1)){
x.a[i,t] <- 2
} #t
} #i
x <- rbind(x.j, x.a)
stan_data <- list(y = CH,
nind = dim(CH)[1],
n_occasions = dim(CH)[2],
x = x,
max_age = max(x))
## Initial values
inits <- function(){list(beta = runif(2, 0, 1),
mean_p = runif(1, 0, 1))}
## Parameters monitored
params <- c("beta", "mean_p")
# MCMC settings
ni <- 2000
nt <- 1
nb <- 1000
nc <- 3
## Stan
model <- stan_model(model_code = cjs_age_code)
cjs_age <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
7.8. Immediate trap response in recapture probability
cjs_trap_code <- "
functions {
int first_capture(int[] y_i) {
for (k in 1:size(y_i))
if (y_i[k])
return k;
return 0;
}
int last_capture(int[] y_i) {
for (k_rev in 0:(size(y_i) - 1)) {
int k;
k <- size(y_i) - k_rev;
if (y_i[k])
return k;
}
return 0;
}
matrix prob_uncaptured(int nind, int n_occasions,
matrix p, matrix phi) {
matrix[nind, n_occasions] chi;
for (i in 1:nind) {
chi[i, n_occasions] <- 1.0;
for (t in 1:(n_occasions - 1)) {
int t_curr;
int t_next;
t_curr <- n_occasions - t;
t_next <- t_curr + 1;
chi[i, t_curr] <- (1 - phi[i, t_curr]) +
phi[i, t_curr] *
(1 - p[i, t_next - 1]) *
chi[i, t_next];
}
}
return chi;
}
}
data {
int<lower=0> nind;
int<lower=2> n_occasions;
int<lower=0,upper=1> y[nind, n_occasions];
int m[nind, n_occasions - 1];
}
transformed data {
int<lower=0,upper=n_occasions> first[nind];
int<lower=0,upper=n_occasions> last[nind];
for (i in 1:nind)
first[i] <- first_capture(y[i]);
for (i in 1:nind)
last[i] <- last_capture(y[i]);
}
parameters {
real<lower=0,upper=1> mean_phi;
real<lower=0,upper=1> mean_p;
real<lower=0,upper=1> beta[2];
}
transformed parameters {
matrix<lower=0,upper=1>[nind, n_occasions - 1] phi;
matrix<lower=0,upper=1>[nind, n_occasions - 1] p;
matrix<lower=0,upper=1>[nind, n_occasions] chi;
// Constraints
for (i in 1:nind) {
for (t in 1:(first[i] - 1)) {
phi[i, t] <- 0;
p[i, t] <- 0;
}
for (t in first[i]:(n_occasions - 1)) {
phi[i, t] <- mean_phi;
p[i, t] <- beta[m[i, t]];
}
}
chi <- prob_uncaptured(nind, n_occasions, p, phi);
}
model {
// Priors
mean_phi ~ uniform(0, 1); // Prior for mean survival
beta ~ uniform(0, 1); // Prior for mean recapture
// Likelihood
for (i in 1:nind) {
if (first[i] > 0) {
for (t in (first[i] + 1):last[i]) {
1 ~ bernoulli(phi[i, t - 1]);
y[i, t] ~ bernoulli(p[i, t - 1]);
}
1 ~ bernoulli(chi[i, last[i]]);
}
}
}
"
## Bundle data
stan_data <- list(y = CH,
nind = dim(CH)[1],
n_occasions = dim(CH)[2],
m = m)
## Initial values
inits <- function(){list(mean_phi = runif(1, 0, 1),
beta = runif(2, 0, 1))}
# Parameters monitored
params <- c("mean_phi", "beta")
# MCMC settings
ni <- 3000
nt <- 2
nb <- 1000
nc <- 4
model <- stan_model(model_code = cjs_trap_code)
cjs_trap <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
7.9. Parameter identifiability
cjs_t_t_code <- "
functions {
int first_capture(int[] y_i) {
for (k in 1:size(y_i))
if (y_i[k])
return k;
return 0;
}
int last_capture(int[] y_i) {
for (k_rev in 0:(size(y_i) - 1)) {
int k;
k <- size(y_i) - k_rev;
if (y_i[k])
return k;
}
return 0;
}
matrix prob_uncaptured(int nind, int n_occasions,
matrix p, matrix phi) {
matrix[nind, n_occasions] chi;
for (i in 1:nind) {
chi[i, n_occasions] <- 1.0;
for (t in 1:(n_occasions - 1)) {
int t_curr;
int t_next;
t_curr <- n_occasions - t;
t_next <- t_curr + 1;
chi[i, t_curr] <- (1 - phi[i, t_curr]) +
phi[i, t_curr] *
(1 - p[i, t_next - 1]) *
chi[i, t_next];
}
}
return chi;
}
}
data {
int<lower=0> nind;
int<lower=2> n_occasions;
int<lower=0,upper=1> y[nind, n_occasions];
}
transformed data {
int<lower=0,upper=n_occasions> first[nind];
int<lower=0,upper=n_occasions> last[nind];
for (i in 1:nind)
first[i] <- first_capture(y[i]);
for (i in 1:nind)
last[i] <- last_capture(y[i]);
}
parameters {
real<lower=0,upper=1> phi_t[n_occasions - 1];
real<lower=0,upper=1> p_t[n_occasions - 1];
}
transformed parameters {
matrix<lower=0,upper=1>[nind, n_occasions - 1] phi;
matrix<lower=0,upper=1>[nind, n_occasions - 1] p;
matrix<lower=0,upper=1>[nind, n_occasions] chi;
// Constraints
for (i in 1:nind) {
for (t in 1:(first[i] - 1)) {
phi[i, t] <- 0;
p[i, t] <- 0;
}
for (t in first[i]:(n_occasions - 1)) {
phi[i, t] <- phi_t[t];
p[i, t] <- p_t[t];
}
}
chi <- prob_uncaptured(nind, n_occasions, p, phi);
}
model {
// Priors
phi_t ~ uniform(0, 1); // Prior for mean survival
p_t ~ uniform(0, 1); // Prior for mean recapture
// Likelihood
for (i in 1:nind) {
if (first[i] > 0) {
for (t in (first[i] + 1):last[i]) {
1 ~ bernoulli(phi[i, t - 1]);
y[i, t] ~ bernoulli(p[i, t - 1]);
}
1 ~ bernoulli(chi[i, last[i]]);
}
}
}
"
## Bundle data
stan_data <- list(y = CH,
nind = dim(CH)[1],
n_occasions = dim(CH)[2])
## Parameters monitored
params <- c("phi_t", "p_t")
## MCMC settings
ni <- 2000
nt <- 1
nb <- 1000
nc <- 3
## Initial values
inits <- lapply(1:nc, function(i) {list(phi_t = runif((dim(CH)[2]-1), 0, 1),
p_t = runif((dim(CH)[2]-1), 0, 1))})
## Stan
model <- stan_model(model_code = cjs_t_t_code)
cjs_t_t <- sampling(model,
data = stan_data, init = inits, pars = params,
chains = nc, thin = nt, iter = ni, warmup = nb,
seed = 1,
open_progress = FALSE)
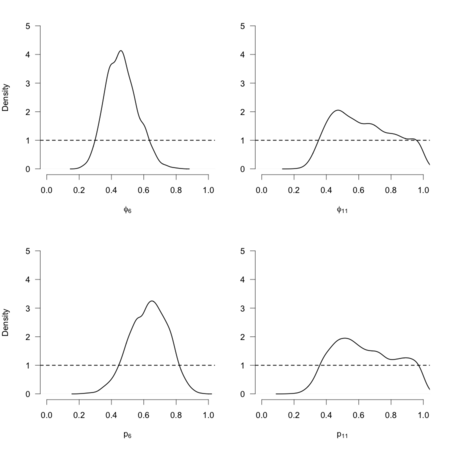
IdentifiabilityがないのはStanで計算させてもおなじ。
タグ:STAn





コメント 0