MCMC: tauの事前分布 [統計]
Carlin & Louis "Bayesian Methods for Data Analysis (3rd ed.)"の45ページ以降で、分散の逆数tauにあたえる事前分布が論じられている。Gamma(ε, ε)という事前分布を使った場合、選択したεにたいして事後分布が鋭敏になる可能性があるということである。
で、例題があったのでためしてみた。model1ではtau ~ dgamma(0.1, 0.1)、model2ではtau ~ dgamma(1.0E-3, 1.0E-3)、model3ではsigma ~ dunif(0.01, 100)としている。
結果
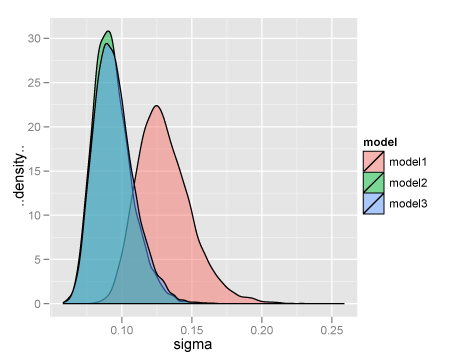
sigmaの事後分布をならべてみる。
ちなみに。
で、例題があったのでためしてみた。model1ではtau ~ dgamma(0.1, 0.1)、model2ではtau ~ dgamma(1.0E-3, 1.0E-3)、model3ではsigma ~ dunif(0.01, 100)としている。
library(R2WinBUGS)
WINE <- "/Applications/Darwine/Wine.bundle/Contents/bin/wine"
WINEPATH <- "/Applications/Darwine/Wine.bundle/Contents/bin/winepath"
model1 <- function() {
for (i in 1:n) {
logage[i] <- log(x[i])
Y[i] ~ dnorm(mu[i], tau)
mu[i] <- beta0 + beta1 * (logage[i] - mean(logage[]))
}
beta0 ~ dflat()
beta1 ~ dflat()
tau ~ dgamma(0.1, 0.1)
sigma <- 1/sqrt(tau)
}
write.model(model1, "model1.bug")
model2 <- function() {
for (i in 1:n) {
logage[i] <- log(x[i])
Y[i] ~ dnorm(mu[i], tau)
mu[i] <- beta0 + beta1 * (logage[i] - mean(logage[]))
}
beta0 ~ dflat()
beta1 ~ dflat()
tau ~ dgamma(1.0E-3, 1.0E-3)
sigma <- 1/sqrt(tau)
}
write.model(model2, "model2.bug")
model3 <- function() {
for (i in 1:n) {
logage[i] <- log(x[i])
Y[i] ~ dnorm(mu[i], tau)
mu[i] <- beta0 + beta1 * (logage[i] - mean(logage[]))
}
beta0 ~ dflat()
beta1 ~ dflat()
tau <- 1/(sigma * sigma)
sigma ~ dunif(0.01, 100)
}
write.model(model3, "model3.bug")
data <- list(
x = c( 1.0, 1.5, 1.5, 1.5, 2.5, 4.0, 5.0, 5.0, 7.0,
8.0, 8.5, 9.0, 9.5, 9.5, 10.0, 12.0, 12.0, 13.0,
13.0, 14.5, 15.5, 15.5, 16.5, 17.0, 22.5, 29.0, 31.5),
Y = c(1.80, 1.85, 1.87, 1.77, 2.02, 2.27, 2.15, 2.26, 2.47,
2.19, 2.26, 2.40, 2.39, 2.41, 2.50, 2.32, 2.32, 2.43,
2.47, 2.56, 2.65, 2.47, 2.64, 2.56, 2.70, 2.72, 2.57),
n = 27)
init1 <- list(beta0 = 0, beta1 = 1, tau = 1)
init3 <- list(beta0 = 0, beta1 = 1, sigma = 1)
params <- c("beta0", "beta1", "sigma")
post1 <- bugs(data = data, inits = list(init1, init1, init1),
parameters.to.save = params,
model.file = "model1.bug",
n.chains = 3, n.iter = 31000, n.burnin = 1000,
n.thin = 10, debug = FALSE,
working.directory = getwd(),
WINE = WINE, WINEPATH = WINEPATH)
post2 <- bugs(data = data, inits = list(init1, init1, init1),
parameters.to.save = params,
model.file = "model2.bug",
n.chains = 3, n.iter = 31000, n.burnin = 1000,
n.thin = 10, debug = FALSE,
working.directory = getwd(),
WINE = WINE, WINEPATH = WINEPATH)
post3 <- bugs(data = data, inits = list(init3, init3, init3),
parameters.to.save = params,
model.file = "model3.bug",
n.chains = 3, n.iter = 31000, n.burnin = 1000,
n.thin = 10, debug = FALSE,
working.directory = getwd(),
WINE = WINE, WINEPATH = WINEPATH)
結果
> print(post1, digits = 3)
Inference for Bugs model at "model1.bug", fit using WinBUGS,
3 chains, each with 31000 iterations (first 1000 discarded), n.thin = 10
n.sims = 9000 iterations saved
mean sd 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta0 2.334 0.025 2.285 2.318 2.334 2.351 2.383 1.001 9000
beta1 0.277 0.028 0.222 0.259 0.277 0.295 0.330 1.002 2400
sigma 0.131 0.019 0.099 0.117 0.128 0.142 0.174 1.001 9000
deviance -46.295 4.800 -53.500 -49.780 -47.025 -43.540 -35.110 1.001 9000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = Dbar-Dhat)
pD = 3.1 and DIC = -43.2
DIC is an estimate of expected predictive error (lower deviance is better).
> print(post2, digits = 3)
Inference for Bugs model at "model2.bug", fit using WinBUGS,
3 chains, each with 31000 iterations (first 1000 discarded), n.thin = 10
n.sims = 9000 iterations saved
mean sd 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta0 2.334 0.018 2.299 2.322 2.334 2.346 2.370 1.001 9000
beta1 0.277 0.020 0.239 0.264 0.277 0.290 0.316 1.001 7300
sigma 0.093 0.014 0.071 0.084 0.092 0.101 0.125 1.001 6200
deviance -52.351 2.596 -55.300 -54.230 -53.020 -51.200 -45.620 1.001 9000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = Dbar-Dhat)
pD = 3.1 and DIC = -49.3
DIC is an estimate of expected predictive error (lower deviance is better).
> print(post3, digits = 3)
Inference for Bugs model at "model3.bug", fit using WinBUGS,
3 chains, each with 31000 iterations (first 1000 discarded), n.thin = 10
n.sims = 9000 iterations saved
mean sd 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta0 2.334 0.018 2.298 2.322 2.334 2.346 2.369 1.001 6000
beta1 0.277 0.020 0.238 0.264 0.277 0.290 0.317 1.001 9000
sigma 0.095 0.014 0.072 0.084 0.093 0.103 0.128 1.001 9000
deviance -52.245 2.621 -55.270 -54.190 -52.920 -50.970 -45.470 1.001 8400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = Dbar-Dhat)
pD = 2.9 and DIC = -49.4
DIC is an estimate of expected predictive error (lower deviance is better).
sigmaの事後分布をならべてみる。

ちなみに。
> summary(lm(data$Y ~ log(data$x)))
Call:
lm(formula = data$Y ~ log(data$x))
Residuals:
Min 1Q Median 3Q Max
-0.148441 -0.054890 0.004223 0.054358 0.168681
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.76166 0.04250 41.45 < 2e-16 ***
log(data$x) 0.27733 0.01881 14.75 7.7e-14 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.08994 on 25 degrees of freedom
Multiple R-squared: 0.8969, Adjusted R-squared: 0.8928
F-statistic: 217.5 on 1 and 25 DF, p-value: 7.707e-14
タグ:MCMC





コメント 0