Stan: 欠測値の推定 [統計]
Stanのマニュアルを見ながら、欠測値の推定をやってみたのでメモ。
下のようなデータがあったとする。
[1,] -3.0150087 0.8224304 -3.5474533 [2,] -2.0796367 1.0398335 -1.4325317 [3,] -2.2329870 1.1210478 -0.9401105 [4,] -2.8172679 -1.5561365 -10.7433671 [5,] -1.2279092 0.5832502 -0.6014049 [6,] -2.1656119 1.5512307 0.3979848 [7,] -1.0271256 1.2240323 1.4461063 [8,] -0.2834660 1.1681719 3.3905226 [9,] -1.7447630 2.0518301 3.1254418 [10,] -1.6334189 1.3834487 0.6055146 [11,] -0.8192108 2.2543968 5.4048861 [12,] -1.3568079 1.6684795 1.3956139 [13,] -0.7046781 1.7936807 3.1889774 [14,] -1.8120819 3.2153371 4.3616880 [15,] -0.4087949 3.8920564 10.9359402 [16,] -2.0551791 2.0741935 1.9299091 [17,] -1.1615289 3.7516962 7.7151888 [18,] -1.8406299 1.7685126 1.7925101 [19,] -1.3740456 2.5434525 4.5620398 [20,] -1.3664153 1.0109986 1.2107255 [21,] -1.3189724 2.3155315 3.3852880 [22,] -2.6820334 4.4423275 8.2837741 [23,] -2.7232567 2.5496929 2.7953275 [24,] -0.3264740 1.9707566 5.1704778 [25,] -2.5957556 1.1692166 -2.3269244 [26,] -0.8401562 3.2464344 8.2907162 [27,] -1.8825776 0.6242447 -1.4520879 [28,] -1.7407786 1.6688916 1.5948284 [29,] -1.6176379 2.9809115 5.5199313 [30,] -2.7114817 4.1907706 7.1706177
これが、下のようにいくつかが欠測になったとする。
x1 x2 y
[1,] -3.0150087 0.8224304 -3.5474533
[2,] NA 1.0398335 -1.4325317
[3,] -2.2329870 1.1210478 -0.9401105
[4,] -2.8172679 -1.5561365 -10.7433671
[5,] -1.2279092 0.5832502 -0.6014049
[6,] -2.1656119 1.5512307 0.3979848
[7,] -1.0271256 1.2240323 1.4461063
[8,] -0.2834660 1.1681719 3.3905226
[9,] -1.7447630 2.0518301 3.1254418
[10,] NA NA 0.6055146
[11,] -0.8192108 2.2543968 5.4048861
[12,] -1.3568079 1.6684795 1.3956139
[13,] NA 1.7936807 3.1889774
[14,] NA 3.2153371 4.3616880
[15,] -0.4087949 NA 10.9359402
[16,] -2.0551791 2.0741935 1.9299091
[17,] -1.1615289 3.7516962 7.7151888
[18,] -1.8406299 1.7685126 1.7925101
[19,] NA 2.5434525 4.5620398
[20,] -1.3664153 NA 1.2107255
[21,] -1.3189724 NA 3.3852880
[22,] -2.6820334 4.4423275 8.2837741
[23,] NA 2.5496929 2.7953275
[24,] -0.3264740 NA 5.1704778
[25,] -2.5957556 1.1692166 -2.3269244
[26,] NA 3.2464344 8.2907162
[27,] -1.8825776 0.6242447 -1.4520879
[28,] -1.7407786 1.6688916 1.5948284
[29,] -1.6176379 2.9809115 5.5199313
[30,] -2.7114817 4.1907706 7.1706177
このようなデータで、欠測部分をStanで推定してみる。Reference Manual の 7.1節に例題があるが、Stanでは欠測値を扱えないので、欠測値の部分をパラメーターにして計算する。RとStanのコードは以下のとおり。x1_missとx2_missの事前分布を指定しないと収束しない。
# Data
set.seed(17)
N <- 30
x1 <- rnorm(N, -2, 1)
x2 <- rnorm(N, 2, 1)
y <- 2 * x1 + 3 * x2 + rnorm(N, 0, 0.5)
# save original values
ox1 <- x1
ox2 <- x2
oy <- y
# Missing data
x1[sample(N, 7)] <- NA
x2[sample(N, 5)] <- NA
print(cbind(x1, x2, y))
## sort
idx0 <- (1:N)[!is.na(x1) & !is.na(x2)]
idx1 <- (1:N)[!is.na(x1) & is.na(x2)]
idx2 <- (1:N)[is.na(x1) & !is.na(x2)]
idx3 <- (1:N)[is.na(x1) & is.na(x2)]
x1 <- x1[c(idx0, idx1, idx2, idx3)]
x2 <- x2[c(idx0, idx1, idx2, idx3)]
y <- y[c(idx0, idx1, idx2, idx3)]
ox1 <- ox1[c(idx0, idx1, idx2, idx3)]
ox2 <- ox2[c(idx0, idx1, idx2, idx3)]
oy <- oy[c(idx0, idx1, idx2, idx3)]
print(cbind(x1, x2, y))
## Stan
library(rstan)
library(parallel)
stancode <- "
data {
int<lower=0> N;
int<lower=0> M0;
int<lower=0> M1;
int<lower=0> M2;
real x1[M0 + M1];
real x2[M0 + M2];
real y[N];
}
transformed data {
int M3;
M3 <- N - M0 - M1 - M2;
}
parameters {
real x1_miss[M2 + M3];
real x2_miss[M1 + M3];
real beta[3];
real<lower=0> sigma;
}
model {
for (i in 1:M0) {
y[i] ~ normal(beta[1] +
beta[2] * x1[i] +
beta[3] * x2[i],
sigma);
}
for (i in (M0 + 1):(M0 + M1)) {
y[i] ~ normal(beta[1] +
beta[2] * x1[i] +
beta[3] * x2_miss[i - M0],
sigma);
}
for (i in (M0 + 1):(M0 + M2)) {
y[M1 + i] ~ normal(beta[1] +
beta[2] * x1_miss[i - M0] +
beta[3] * x2[i],
sigma);
}
for (i in 1:M3) {
y[M0 + M1 + M2 + i] ~ normal(beta[1] +
beta[2] * x1_miss[M2 + i] +
beta[3] * x2_miss[M1 + i],
sigma);
}
x1_miss ~ normal(mean(x1), sd(x1));
x2_miss ~ normal(mean(x2), sd(x2));
}
"
model <- stan_model(model_code = stancode)
data <- list(N = N,
M0 = length(idx0),
M1 = length(idx1),
M2 = length(idx2),
x1 = x1[!is.na(x1)],
x2 = x2[!is.na(x2)],
y = y)
seeds <- c(1, 2, 3, 5)
sflist <- mclapply(1:4,
function(i) {
sampling(model,
data = data, seed = seeds[i],
chains = 1, iter = 2000)},
mc.cores = 4)
fit <- sflist2stanfit(sflist)
##
library(ggplot2)
samp <- extract(fit, pars = c("x1_miss", "x2_miss"))
q.x1_miss <- apply(samp$x1_miss, 2, quantile, probs = c(0.025, 0.5, 0.975))
q.x2_miss <- apply(samp$x2_miss, 2, quantile, probs = c(0.025, 0.5, 0.975))
df <- data.frame(var = c(rep("x1_miss", ncol(q.x1_miss)),
rep("x2_miss", ncol(q.x2_miss))),
x = as.factor(c(1:ncol(q.x1_miss),
1:ncol(q.x2_miss))),
orig = c(ox1[is.na(x1)], ox2[is.na(x2)]),
low = c(q.x1_miss[1, ], q.x2_miss[1, ]),
med = c(q.x1_miss[2, ], q.x2_miss[2, ]),
high = c(q.x1_miss[3, ], q.x2_miss[3, ]))
p <- ggplot(df) +
geom_pointrange(aes(x = x, y = med, ymax = high, ymin = low),
colour = "red", size = 1, alpha = 0.8) +
geom_point(aes(x = x, y = orig), shape = 3, size = 4,
colour = "black", alpha = 1) +
xlab("x") + ylab("value") +
facet_wrap(~ var, scales = "free_x")
print(p)
結果
Inference for Stan model: stancode.
4 chains, each with iter=2000; warmup=1000; thin=1;
post-warmup draws per chain=1000, total post-warmup draws=4000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
x1_miss[1] -2.19 0.00 0.27 -2.71 -2.37 -2.19 -2.01 -1.65 4000 1
x1_miss[2] -1.12 0.00 0.26 -1.62 -1.29 -1.12 -0.96 -0.62 2912 1
x1_miss[3] -2.50 0.00 0.27 -3.03 -2.67 -2.50 -2.34 -1.97 4000 1
x1_miss[4] -1.52 0.00 0.26 -2.03 -1.68 -1.52 -1.36 -1.00 4000 1
x1_miss[5] -2.33 0.00 0.26 -2.84 -2.50 -2.32 -2.15 -1.81 4000 1
x1_miss[6] -0.79 0.00 0.28 -1.35 -0.96 -0.78 -0.61 -0.24 3839 1
x1_miss[7] -1.80 0.02 0.76 -3.32 -2.30 -1.81 -1.30 -0.32 2520 1
x2_miss[1] 3.91 0.00 0.21 3.51 3.78 3.91 4.05 4.33 3482 1
x2_miss[2] 1.34 0.00 0.18 0.97 1.22 1.34 1.46 1.68 4000 1
x2_miss[3] 2.03 0.00 0.18 1.67 1.91 2.02 2.14 2.40 4000 1
x2_miss[4] 1.95 0.00 0.20 1.56 1.83 1.95 2.08 2.34 3049 1
x2_miss[5] 1.43 0.01 0.53 0.38 1.08 1.44 1.78 2.47 2391 1
beta[1] 0.00 0.01 0.36 -0.72 -0.23 -0.01 0.23 0.70 1946 1
beta[2] 2.01 0.00 0.16 1.70 1.91 2.02 2.11 2.34 2209 1
beta[3] 2.98 0.00 0.09 2.81 2.92 2.98 3.04 3.15 3013 1
sigma 0.52 0.00 0.10 0.37 0.45 0.51 0.58 0.76 1383 1
lp__ 0.65 0.14 3.86 -8.15 -1.71 1.03 3.49 6.76 795 1
Samples were drawn using NUTS(diag_e) at Sat Mar 7 14:59:25 2015.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
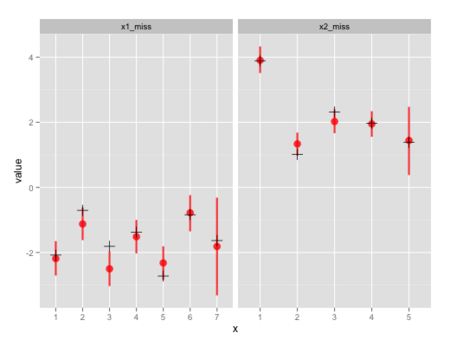
赤の点と線が事後分布の中央値および95%信用区間、黒の十字が元のデータ。
だいたいうまく推定できているが、x1とx2の両方が欠測になったx1_miss[7]とx2_miss[5]では信用区間が広くなっている。
タグ:RStan





コメント 0