glmmAK [統計]
CRANにglmmAKというパッケージが登録されていた。AKというのは作者のArnošt Komárek氏の名前からとったものか。
This package implements several generalized linear (mixed) models (GLMM) where for random effects either a conventional normal distribution is assumed or a flexible model based on penalized smoothing is used for the random effect distribution. We call the second class of models as penalized Gaussian mixture (PGM) or shortly G-spline.
通常のGLMMではrandom effectが正規分布することを仮定するが、このパッケージではpenalized Gaussian mixture (PGM)とすることができるということのようだ。で、PGMというのはG-スプラインということか。このあたり勉強不足なのでよく分からないのだが。
通常のGLMMのrandam effect項
![]()
PGMのrandom effect項
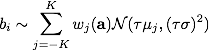
MCMCで推定をおこなうようになっている。
付属の例題(ex-Toenail)をやってみる。1) PGM GLMM, not hierarchically centered, 2) PGM GLMM, hierarchically centered, 3) Normal GLMM, not hierarchically centered, 4) Normal GLMM, hierarchically centeredの4つを比較しているが、とりあえず1)だけをやってみる。
まずはいろいろ準備。
library(glmmAK)
root <- paste(Sys.getenv("HOME"), "Documents/RData/", sep="/")
setwd(root)
data(toenail)
# 事前分布
prior.fixed <- list(mean = 0, var = 10000)
prior.gspline.Slice <- list(K = 15, delta = 0.3,
sigma = 0.2, CARorder = 3,
Ldistrib = "gamma", Lequal = FALSE, Lshape = 1,
LinvScale = 0.005,
AtypeUpdate = "slice")
prior.gspline.Block <- list(K = 15, delta = 0.3,
sigma = 0.2, CARorder = 3,
Ldistrib = "gamma", Lequal = FALSE, Lshape = 1,
LinvScale = 0.005,
AtypeUpdate = "block")
prior.random.gspl.nhc <- list(Ddistrib = "gamma",
Dshape = 1, DinvScale = 0.005)
# 結果のファイルを収めるpathの準備
if (!("chToenail" %in% dir(root))) dir.create(paste(root, "chToenail",
sep = ""))
dirNames <- c("PGM_nhc")
dirPaths <- paste(root, "chToenail/", dirNames, "/", sep = "")
for (i in 1:length(dirPaths)) {
if (!(dirNames[i] %in% dir(paste(root, "chToenail", sep = ""))))
dir.create(dirPaths[i])
}
names(dirPaths) <- c("PGM_nhc")
# 共変量
iXmat <- data.frame(trt = toenail$trt,
time = toenail$time,
trt.time = toenail$trt * toenail$time)
# MCMCの回数
# niter × nthinが繰り返し回数となる
# 例題ではnthin = 100, nwrite = 100となっているが、それぞれ10に
nsimul <- list(niter = 4000, nthin = 10, nburn = 2000, nwrite = 10)
MCMCを実行。
fit.PGM.nhc <- cumlogitRE(y = toenail$infect, x = iXmat,
cluster = toenail$idnr, intcpt.random = TRUE,
hierar.center = FALSE,
C = 1, drandom = "gspline", prior.fixed = prior.fixed,
prior.random = prior.random.gspl.nhc,
prior.gspline = prior.gspline.Slice,
nsimul = nsimul, store = list(prob = FALSE, b = TRUE),
dir = dirPaths["PGM_nhc"])
事後分布を読み込む。
chPGM.nhc <- glmmAK.files2coda(dir = dirPaths["PGM_nhc"],
drandom = "gspline",
skip = 0)
iters <- scanFH(paste(dirPaths["PGM_nhc"], "iteration.sim", sep = ""))
betaF <- scanFH(paste(dirPaths["PGM_nhc"], "betaF.sim", sep = ""))
betaRadj <- scanFH(paste(dirPaths["PGM_nhc"], "betaRadj.sim",
sep = ""))
varRadj <- scanFH(paste(dirPaths["PGM_nhc"], "varRadj.sim", sep = ""))
chPGM.nhc <- mcmc(data.frame(Trt = betaF[, "trt"],
Time = betaF[, "time"],
Trt.Time = betaF[, "trt.time"],
Meanb = betaRadj[, "(Intercept)"],
SDb = sqrt(varRadj[, "varR.1"])),
start = iters[1, 1])
rm(list = c("iters", "betaF", "betaRadj", "varRadj"))
結果。
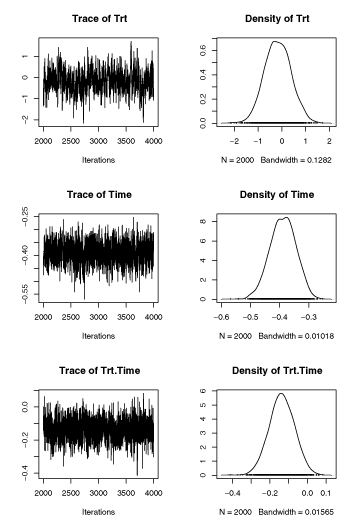
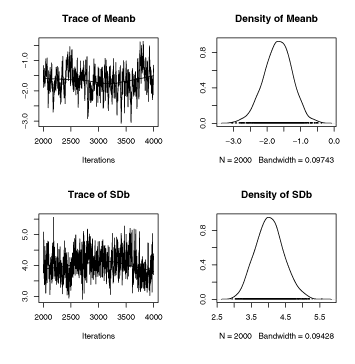
summary(chPGM.nhc)
Iterations = 2001:4000
Thinning interval = 1
Number of chains = 1
Sample size per chain = 2000
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
Trt -0.1707 0.55305 0.0123667 0.038592
Time -0.3888 0.04393 0.0009823 0.001471
Trt.Time -0.1364 0.06750 0.0015093 0.002118
Meanb -1.6728 0.42032 0.0093987 0.048364
SDb 4.0355 0.40676 0.0090954 0.032822
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
Trt -1.2368 -0.5494 -0.1756 0.20957 0.914094
Time -0.4759 -0.4183 -0.3876 -0.35809 -0.308134
Trt.Time -0.2688 -0.1810 -0.1373 -0.08957 -0.005997
Meanb -2.5440 -1.9513 -1.6625 -1.38506 -0.897988
SDb 3.2792 3.7500 4.0270 4.29516 4.873613
plot(chPGM.nhc)
lmerと比較してみる。
library(lme4)
glmm.lmer <- lmer(infect ~ trt + time + trt:time + (1|idnr),
family=binomial, data=toenail, method="Laplace")
Generalized linear mixed model fit using Laplace
Formula: infect ~ trt + time + trt:time + (1 | idnr)
Data: toenail
Family: binomial(logit link)
AIC BIC logLik deviance
1266 1293 -627.8 1256
Random effects:
Groups Name Variance Std.Dev.
idnr (Intercept) 20.867 4.568
number of obs: 1908, groups: idnr, 294
Estimated scale (compare to 1 ) 1.623704
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.51388 0.46275 -5.432 5.56e-08 ***
trt -0.30664 0.66293 -0.463 0.6437
time -0.40044 0.04520 -8.859 < 2e-16 ***
trt:time -0.13629 0.07361 -1.851 0.0641 .
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) trt time
trt -0.698
time -0.277 0.193
trt:time 0.170 -0.278 -0.614





コメント 0